With more than 20% of infant deaths and about 40% of prenatal deaths each year, congenital heart diseases (CHD) are the most common congenital disorders in newborns, thus representing a growing burden for health care systems.1 Furthermore, an increasing number of children affected by even the most complex forms of CHD are surviving to adulthood, exceeding the pediatric CHD cohort and thus requiring lifelong specialized cardiac care.1 As a consequence, there is an urgent need to better understand the genetic and molecular mechanisms associated with the disease in order to establish new preventive strategies and treatments.
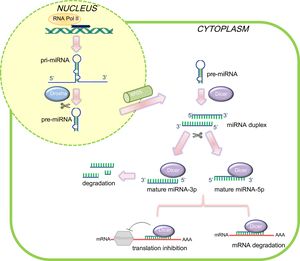
Several decades of intensive research have revealed the highly complicated regulatory networks governing cardiovascular development, underscoring the importance of genetic factors as well as environmental cues in the pathogenesis of CHD. Recently, a novel mechanism involving posttranscriptional regulation by microRNAs (miRNAs) has emerged as a central regulator of embryonic heart development.2 MicroRNAs are a class of small and evolutionarily preserved regulatory RNAs, approximately 20 to 26 nucleotides in length at a mature stage. These molecules act at a posttranscriptional level primarily by an imperfect base pairing with the mRNA target in a sequence-specific manner, resulting in translational repression and gene silencing (Figure 1).3
Schematic representation of microRNAs (miRNAs) biogenesis and mode of action in mammalian cells. In the canonical miRNA biogenesis pathway, pri-miRNAs are transcribed in the nucleus by RNA polymerase II and processed by Drosha into a shorter precursor of ∼65 nucleotides in length (pre-miRNA), which is subsequently exported out of the nucleus. In the cytoplasm, Dicer recognizes and cleaves the pre-miRNA molecules, producing unstable ∼22 nucleotide miRNA duplex structures. Typically, the arm with the freer 5’end is preferentially incorporated into RISC in order to direct mRNA target silencing. Nevertheless, the biological function of the other strand is still under debate. The complementary pairing between mRNA and mature miRNA determines the gene silencing mechanism. mRNA, messenger RNA.
From reports published to date, various forms of CHDs have been associated with altered expression of specific miRNAs and associated target genes, thus suggesting that a proper regulation of miRNA expression levels during cardiogenesis might be crucial for CHD prevention. During the last few decades, deciphering the significance of miRNA-mediated posttranscriptional regulation in CHD pathology has benefited from extensive model organism studies that have provided new insights into heart development. Specifically, earlier studies have examined the tissue-specific deletion of enzymes essential for miRNA biogenesis (eg, Dicer, Drosha, Ago2, DGCR8) in mice and zebrafish models. Indeed, knock-out models resulted in early embryonic lethality due to general growth arrest and defects in heart development, demonstrating the importance of miRNAs in the cardiovascular system.3 Such roles have been further confirmed by gain- or loss-of-function experiments, indicating that a fine balance in miRNA abundance is fundamental to maintaining the cardiac homeostasis and that a deviation from such a balance may play a substantial role in cardiovascular disease.4
Cardiac muscle is enriched with numerous miRNAs, such as miR-1, miR-133, miR-499 and miR-208, which are abundantly, but not exclusively, expressed in the cardiac tissue. Studies on the developing murine and Xenopus laevis heart demonstrated that misexpression of such muscle-specific microRNAs altered cardiac tissue formation, leading to a thin ventricular wall and early embryonic lethality due to severe cardiac malformations.5 Recent high-throughput technologies combining bioinformatic and molecular analysis have definitively established the power of such miRNAs to modulate, and even control, fundamental cardiac transcription factors, such as Gata4, MEF2C, Tbx1, cardiac myosin heavy chain genes and SRF, whose deregulation have been previously associated with the development of CHD.4
Additional ubiquitously expressed miRNAs have been functionally analyzed, including miR-17-92, miR-195, miR-196a and miR-363, and have been demonstrated to play a role in controlling myocardial differentiation during mammalian embryogenesis. Indeed, additional in vivo evidence has demonstrated that altered expression of such miRNAs may induce conotruncal heart malformations and/or septal defects in distinct animal models, probably by repressing key cardiac progenitor genes, such as Isl1, Tbx1, Hoxb8, and Hand1.6
In this regard, it is intriguing that miRNAs may act through feedback loops to regulate their own expression and that of their target mRNAs. This is the case of miR-1 in muscle cells, which can modify the expression of histone deacetylase 4 (HDAC4), a repressor of MEF2 transcription factor, which, in turn, activates miR-1 and other target mRNAs.4,7 This fine regulation feedback loop might play a key role in cellular proliferation and differentiation, probably contributing to structural and/or functional cardiac-related disorders.
Altogether, the findings of in vivo and in vitro studies have definitively created the optimal background for human studies, characterizing a wide range of miRNAs that might play a role in the development of CHD in humans when dysregulated.
Notably, deregulated miRNA expression patterns in humans have been predominantly documented in cyanotic diseases and ventricular septal defects, highlighting the presence of an altered miRNA profile, mostly in the myocardial human tissue. Specifically, multiple differentially-expressed miRNAs, including miR-1, miR-206, miR-421, miR-940, miR-181c and miR-138, have been reported to be associated with tetralogy of Fallot and septal defects, the most common forms of CHD.8,9 Additionally, an atypical miRNA expression profile has been recently recognized in the right ventricular myocardium of patients with hypoplastic left heart syndrome patients, affecting the expression of several target genes important for proper heart development.10
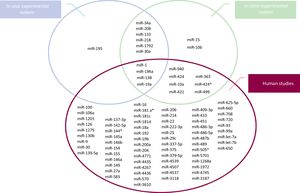
Figure 2 summarizes specific aberrantmiRNAexpression from experimental and human investigations. Although the findings of all these studies have provided a rationale to investigate further into the potential of miRNAs during heart development, profiling of such miRNAs in human CHD carries some intrinsic challenges. Indeed, the wide heterogeneity of miRNA expression deduced from all these studies reflects the complex etiology of CHD and the high genetic heterogeneity between CHD patients. Additionally, the identified microRNAs can be biased by the subtype of CHD, the small sample size, and the specimen used for the analysis (eg, cell lines, heart tissue, or blood). Fortunately, many of these limitations have now been overcome by the recent advent of high-throughput techniques, and a wide range of approaches focused on sensitivity and specificity are now available for miRNA detection.
The growing interest in the miRNA profile and its association with different types of CHD has laid the foundation to look for a role of circulating miRNAs as potential diagnostic and prognostic biomarkers. Indeed, due to their high stability in blood/serum, miRNAs released into the circulation could be potentially used as noninvasive biomarkers for prenatal screening and prognosis evaluation. To date, only a few pilot studies have explored differentially expressed miRNAs in the blood/serum of CHD patients, highlighting the need to further explore their potential application in the clinical setting.9 Interestingly, some data have correlated the expression of placental miRNAs in the sera of pregnant women with CHD, suggesting new possibilities in terms of noninvasive prenatal tests of CHD.11
Another hypothesis has been recently postulated regarding the effect of genetic variants within miRNA genes (ie, pri-miRNAs, pre-miRNAs and mature miRNAs) on the biogenesis and function of specific microRNAs.12 Indeed, gain- and loss-of-function miRNA's single nucleotide polymorphisms (SNPs) may affect the processing and the subsequent maturation of mRNAs as well as the target selection of miRNAs, influencing the risk and/or prognosis of a variety of human diseases, including several cardiac disorders.
Regarding CHD susceptibility, rs11614913 SNP in miR-196a2, located at the pre-miRNA sequence, seems to be the most promising SNP, although its biological consequences on heart development remain controversial and require additional studies.13 Nevertheless, few variants in miRNA genes have been associated with CHD risk so far, and the relationship between miRNA SNPs and CHD remains largely unknown. These findings are consistent with the fact that genetic variants within miRNA genes occur at a very low frequency (about 10% in pre-miRNA sequence and less than 1% within the seed region), highlighting the rigidly preserved seed sequence identity both within and between species.
In line with this, the presence of SNPs in the 3’-untranslated region (UTR) of miRNA target genes has attracted much attention and has also been shown to play a role in some forms of CHD. This is the case of the rs3203358 and rs6489956 SNPs, located in the 3’UTR of GATA4 and Tbx5, respectively. These genes encode for 2 of the best-characterized transcription factors that are critical for normal heart development. Indeed, 2 elegant studies have demonstrated a significant effect of these 2 variants on the increased susceptibility of CHD by affecting the binding ability of specific miRNAs.14,15 To date, although undoubtedly relevant, only very few studies have investigated the potential impact of miRNA-related SNPs on CHD risk. Comprehensive and integrative approaches may be useful to better understand the effects of genetic variants on the cell regulatory circuit managed by specific miRNAs, whose deregulation may lead to structural and functional heart malformations. Indeed, investigating which genes and pathways are affected may lead to a deeper understanding of the molecular mechanisms during cardiogenesis under both physiological and pathological conditions, and ultimately highlight novel actionable targets for specific miRNA-based therapies.
Although the study of miRNAs in the pathogenesis of CHD is still relatively new, these molecules hold great promise as crucial players of heart development, offering novel opportunities to further elucidate the wide heterogeneity of CHD and improve its prenatal diagnosis.
CONFLICTS OF INTERESTNone declared.