Cardiovascular disease is a multifactorial, polygenic, complex disease with a substantial environmental component. This makes risk prediction and stratification on an individual level highly challenging. Technological advances and availability of larger, well-conducted studies have resulted in substantial progress defining the impact of common genetic variation on cardiovascular disease progression. The discovery of common genetic variants, including the chromosome 9p21.3 locus as the strongest and most highly replicated independent genetic coronary disease risk factor, has further stimulated interest in genetic testing for cardiac-associated risk variants. Adding to the understanding of the genetics of cardiovascular disease is the growing information available on the role of transcripts or gene expression. Likely, the combination of DNA, RNA, and clinical data will help clarify many of the complexities seen in cardiovascular disease.
Genetic Associations and Cardiovascular Disease
Available human genome-wide association studies have analyzed over 500 000 single nucleotide polymorphisms in large numbers of subjects and genetically defined diverse diseases and risk factors.1-3 However, despite great progress in the diagnosis and treatment of unstable coronary syndromes it is estimated that new and recurrent cardiac events will continue to be a major clinical burden in the foreseeable future.4 Central to the pathogenesis of acute coronary syndromes is the adhesion and activation of platelets leading to thrombus formation and vessel occlusion and the absolute risk of recurrent vascular events among patients taking platelet inhibitors remains high.4 The observation that platelet dependent thrombosis occurs, despite treatment with platelet inhibitors including aspirin and clopidogrel, has generated many additional studies searching for alternative causes of treatment failure. Data suggest that endogenous platelet function is partially genetically determined with a wide inter-individual variability in the platelet activation response.5 Heritable factors also play a major role in platelet function6 although, importantly, genetic variants of platelet receptors have not consistently been shown to correlate with platelet function, alter drug response, or associate consistently with cardiovascular disease in larger studies.6
Transcriptomics in Cardiovascular Disease: The Blood Transcriptome
Our understanding of cardiovascular disease has been assisted by developments in the burgeoning fields of genomics and proteomics. Transcriptomics, specifically, has been previously successfully utilized only in large-scale gene expression profiling in oncological settings. Limited information is available in cardiovascular syndromes due to availability of appropriate tissue that would be relevant and a useful surrogate of disease. In addition, many previous studies in selected vascular disease are limited by small size. A recent study demonstrated that specific peripheral blood transcripts play a role in the pathogenesis of coronary heart disease and its risk factors.7 Using a large community-based cohort, inflammatory transcripts derived from platelets were found to be associated with body mass index.7 Other transcripts, particularly those derived from peripheral blood mononuclear cells, were found to be heritable.7
In addition to the study of the transcript or messenger RNA (mRNA), another area of gene expression that is being directed towards study of blood derived transcripts in cardiovascular disease are micro RNAs (miRNAs); miRNAs have been recently discovered to be small RNAs that negatively regulate gene expression by suppressing protein translation. The expression of many miRNAs may reflect selected diseases8-9 and over 800 human miRNAs have been identified. Bioinformatic predictions suggest that mammalian miRNAs can regulate approximately 30% of all protein-coding genes. Micro RNAs have been found in whole blood and may be a useful way to further define cardiovascular disease.
The Use of Genetic Testing in the Treatment of Coronary Disease
Pharmacogenomics tries to identify specific gene variants that are able to explain the variability in patient response to a given drug. The two specific questions it can answer are: a) can individual genetic characteristics guide medical practice and medication use now; and b) will pharmacogenetics contribute to personalized medicine in the future?
The 2 main categories in which pharmacogenomics can add to the treatment of cardio-thrombotic disease include: a) the use of genomics to understand the variable response in existing drugs and the use to guide therapy; and, b) the use of genomic information to identify new drug targets. While most of the current literature and data are speculative in terms of direct applicability to guiding treatment, there are a few exceptions which will be described below:
Warfarin
Warfarin/coumadin is prescribed to millions of individuals annually for atrial fibrillation, prosthetic heart valves, surgery or a history of vascular thrombosis. Warfarin's use and clinical management are difficult due to a narrow therapeutic index and many other reasons for variability in levels. The combination of genotype and clinical factors (age, weight, and sex) explains approximately 50%-60% of variance in warfarin dose requirements although racial differences exist. Other algorithms have reported predicting up to 70% of interindividual dose variability. However, genetic testing has not been shown to help with the risk for over-anticoagulation and bleeding or anticoagulation control. Using pharmacogenetic and clinical factors can improve the accuracy and efficiency of warfarin dose initiation although not in terms of time in target international normalized ratio.10 Thus, although promising, current routine use of CYP2C9 or VKORC1 genotyping is not fully supported by available literature. Although it should be noted that the U.S. Food and Drug Administration (2007) has added a cautionary note to warfarin labeling stating that "a lower initiation dose should be considered for patients with certain genetic variations in CYP2C9 and VKORC1 enzymes."
Clopidogrel
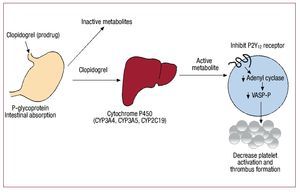
For the platelet P2Y12 receptor inhibitor clopidogrel, there is growing supportive data suggesting that select genetics may impact on both drug responsiveness and utility. However, the responsible genetic variant appears not to be the expected P2Y12 receptor, but cytochrome P450 (CYP), an enzyme responsible for its metabolism (Figure). Clopidogrel is a pro-drug which undergoes liver metabolism by specific CYP enzymes and these genes encode the CYP-dependent oxidative steps. The genes are polymorphic and previous studies have shown that carriers of the specific alleles of CYP2C1911-13 and CYP3A414 have increased platelet function. Increased platelet activity has also been specifically associated with CYP2C19*2 allele which causes loss of function in several different populations including patients after coronary stent placement15 and in clopidogrel treated patients post non-ST elevation myocardial infarction.16 Prasugrel, a P2Y12 inhibitor not hepatically metabolized, does not appear to be affected by P450 isoenzyme variability. In a cohort of over 2200 clopidogrel treated patients presenting with acute myocardial infarction,17 it has been shown that those carrying any two CYP2C19 loss-of-function alleles have a higher rate of cardiac events. Carriers of the ABCB1 variant that modulates clopidogrel absorption also had a modestly increased rate of events. In a study examining genetic variants in CYP genes, clopidogrel plasma concentration, and platelet function in healthy subjects as well as in nearly 1500 patients presenting with an acute coronary syndrome and treated with clopidogrel from the TRITON-TIMI38 trial,18 investigators found that, in healthy subjects, carriers of at least one CYP2C19 loss-of-function allele had decreased active clopidogrel metabolite compared to non-carriers and decreased platelet aggregation. In clopidogrel treated subjects from TRITON-TIMI38, carriers of the loss of function alleles had increased risk of cardiovascular death, myocardial infarction or stroke as compared to non carriers.
Figure 1. Metabolism and mechanism of action of clopidogrel. Clopidogrel is a prodrug that is primarily metabolized into inactive metabolites by esterases. The active metabolites are generated via hepatic cytochrome P450 enzymes and these active metabolites inhibit the adenosine diphosphate P2Y12 receptor decreasing platelet activation and thrombus formation.
In March, 2010, the United States Food and Drug Administration approved a new (updated) label for clopidogrel with a "boxed warning" stating the potential for diminished effectiveness in patients with decreased CYP2C19 function.19 The current updated label no longer specifically advises avoidance of clopidogrel in patients with this polymorphism but states that alternative treatment should be considered. Consistent with the current lack of prospective clinical data using a different therapy or modified dose; clear recommendations for genotyping are not longer clearly stated.
Antiplatelet therapy is associated with increased risk of gastrointestinal bleeding and many patients are also prescribed proton pump inhibitors to prevent these side-effects. Most proton pump inhibitors are predominantly metabolized by CYPP450 enzymes. Consistent with the importance of hepatic enzymes, coadminstration of omeprazole, also metabolized by CYP2C19, has been shown to decrease clopidogrel's platelet inhibitory effect.20 However, the coadministration of proton pump inhibitors is currently contentious as the studies have been inconsistent with some showing no impact on the clinical response to clopidogrel.18 The discrepancy between this finding and the previous reports is unclear and the current recommendations vary between groups and agencies.
Conclusions
The use of genetic testing in cardiovascular disease and therapeutics will continue to grow as larger, better conducted studies are utilized along with high throughput technologies. This includes using: a) candidate gene approaches with select genes/ single nucleotide polymorphisms chosen based on suspicion of association; and b) genome-wide approaches. Studies will include genome scanning that tests association of microsatellite markers located throughout the genome with a phenotype; wide SNP maps across the genome; and wide transcriptomics. With the currently available data, many pivotal questions remain unanswered and would require larger studies showing that therapeutic interventions based on genetic testing will lead to better clinical outcome and measuring genetic markers adds to diagnose disease. In summary, our understanding and the depth of information is rapidly growing and points to a future for genetic/transcript testing in cardiovascular disease.
Correspondence: J. E. Freedman, MD
Boston University School of Medicine,
700 Albany Street, W507 Boston, MA 02118. United States
E-mail: freedmaj@bu.edu